import json
import sys
import requests
import folium
import shapely
import pandas as pd
import matplotlib.pyplot as pltGet timeseries from COGs
Run this notebook
You can launch this notbook using mybinder, by clicking the button below.
Approach
- Using a list of STAC items and a bouding box fetch stats from
/statisticsendpoint - Generate a timeseries plot using statistics from each time step
- Speed up workflow using Dask
The /statistics endoint is provided by the VEDA implementation of titiler. This service provides an API that can be used by websites and programmatically to produce dynamic tiles and data aggretations. For VEDA, it also provides a public interface for the data since the API is publicly accessible even though the underlying data requires AWS authentication or running on the hub.
Titiler is a powerful tool for visualization but has inherent limitations when compared with direct data access. One of these limitations is that the body of any given request must be smaller than 16Kb. This can create issues when the request contains verbose content such as geojson. Just remember that as long as you are on the hub, you always have the ability to access the data directly and do any manipulations within the notebook.
Declare your collection of interest
You can discover available collections the following ways:
- Programmatically: see example in the
list-collections.ipynbnotebook - JSON API: https://openveda.cloud/api/stac/collections
- STAC Browser: https://openveda.cloud
STAC_API_URL = "https://openveda.cloud/api/stac"
RASTER_API_URL = "https://openveda.cloud/api/raster"
collection_id = "no2-monthly"Fetch STAC collection
We will use requests to fetch all the metadata about the collection of interest from STAC.
response = requests.get(f"{STAC_API_URL}/collections/{collection_id}")
assert response.ok, response.text
collection = response.json()
collection{'id': 'no2-monthly',
'type': 'Collection',
'links': [{'rel': 'items',
'type': 'application/geo+json',
'href': 'https://openveda.cloud/api/stac/collections/no2-monthly/items'},
{'rel': 'parent',
'type': 'application/json',
'href': 'https://openveda.cloud/api/stac/'},
{'rel': 'root',
'type': 'application/json',
'href': 'https://openveda.cloud/api/stac/'},
{'rel': 'self',
'type': 'application/json',
'href': 'https://openveda.cloud/api/stac/collections/no2-monthly'},
{'rel': 'http://www.opengis.net/def/rel/ogc/1.0/queryables',
'type': 'application/schema+json',
'title': 'Queryables',
'href': 'https://openveda.cloud/api/stac/collections/no2-monthly/queryables'}],
'title': 'NO₂',
'assets': {'thumbnail': {'href': 'https://thumbnails.openveda.cloud/no2--dataset-cover.jpg',
'type': 'image/jpeg',
'roles': ['thumbnail'],
'title': 'Thumbnail',
'description': 'Photo by [Mick Truyts](https://unsplash.com/photos/x6WQeNYJC1w) (Power plant shooting steam at the sky)'}},
'extent': {'spatial': {'bbox': [[-180.0, -90.0, 180.0, 90.0]]},
'temporal': {'interval': [['2016-01-01T00:00:00+00:00',
'2022-12-31T00:00:00+00:00']]}},
'license': 'MIT',
'renders': {'dashboard': {'bidx': [1],
'title': 'VEDA Dashboard Render Parameters',
'assets': ['cog_default'],
'rescale': [[0, 15000000000000000]],
'resampling': 'bilinear',
'color_formula': 'gamma r 1.05',
'colormap_name': 'rdbu_r'}},
'providers': [{'url': 'https://disc.gsfc.nasa.gov/',
'name': 'NASA Goddard Earth Sciences Data and Information Services Center',
'roles': ['producer', 'processor']},
{'url': 'https://www.earthdata.nasa.gov/dashboard/',
'name': 'NASA VEDA',
'roles': ['host']}],
'summaries': {'datetime': ['2016-01-01T00:00:00Z', '2023-09-30T00:00:00Z']},
'description': 'Darker colors indicate higher nitrogen dioxide (NO₂) levels and more activity. Lighter colors indicate lower levels of NO₂ and less activity. Missing pixels indicate areas of no data most likely associated with cloud cover or snow.',
'item_assets': {'cog_default': {'type': 'image/tiff; application=geotiff; profile=cloud-optimized',
'roles': ['data', 'layer'],
'title': 'Default COG Layer',
'description': 'Cloud optimized default layer to display on map'}},
'stac_version': '1.0.0',
'stac_extensions': ['https://stac-extensions.github.io/item-assets/v1.0.0/schema.json',
'https://stac-extensions.github.io/render/v1.0.0/schema.json'],
'dashboard:is_periodic': True,
'dashboard:time_density': 'month'}Describe the periodic nature of the data
In the collection above we will pay particular attention to the fields that define the periodicity of the data.
collection["dashboard:is_periodic"]Truecollection["dashboard:time_density"]'month'collection["summaries"]{'datetime': ['2016-01-01T00:00:00Z', '2023-09-30T00:00:00Z']}Fetch STAC items
Get the list of all the STAC items within this collection.
response = requests.get(
f"{STAC_API_URL}/collections/{collection_id}/items?limit=100"
)
assert response.ok, response.text
items = response.json()["features"]
len(items)93We can inspect one of these items to get a sense of what metadata is available.
items[0]{'id': 'OMI_trno2_0.10x0.10_202309_Col3_V4.nc',
'bbox': [-180.0, -90.0, 180.0, 90.0],
'type': 'Feature',
'links': [{'rel': 'collection',
'type': 'application/json',
'href': 'https://openveda.cloud/api/stac/collections/no2-monthly'},
{'rel': 'parent',
'type': 'application/json',
'href': 'https://openveda.cloud/api/stac/collections/no2-monthly'},
{'rel': 'root',
'type': 'application/json',
'href': 'https://openveda.cloud/api/stac/'},
{'rel': 'self',
'type': 'application/geo+json',
'href': 'https://openveda.cloud/api/stac/collections/no2-monthly/items/OMI_trno2_0.10x0.10_202309_Col3_V4.nc'},
{'title': 'Map of Item',
'href': 'https://openveda.cloud/api/raster/collections/no2-monthly/items/OMI_trno2_0.10x0.10_202309_Col3_V4.nc/map?bidx=1&assets=cog_default&rescale=0%2C15000000000000000&resampling=bilinear&color_formula=gamma+r+1.05&colormap_name=rdbu_r',
'rel': 'preview',
'type': 'text/html'}],
'assets': {'cog_default': {'href': 's3://veda-data-store/no2-monthly/OMI_trno2_0.10x0.10_202309_Col3_V4.nc.tif',
'type': 'image/tiff; application=geotiff',
'roles': ['data', 'layer'],
'title': 'Default COG Layer',
'proj:bbox': [-180.0, -90.0, 180.0, 90.0],
'proj:epsg': 4326,
'proj:wkt2': 'GEOGCS["WGS 84",DATUM["WGS_1984",SPHEROID["WGS 84",6378137,298.257223563,AUTHORITY["EPSG","7030"]],AUTHORITY["EPSG","6326"]],PRIMEM["Greenwich",0],UNIT["degree",0.0174532925199433,AUTHORITY["EPSG","9122"]],AXIS["Latitude",NORTH],AXIS["Longitude",EAST],AUTHORITY["EPSG","4326"]]',
'proj:shape': [1800, 3600],
'description': 'Cloud optimized default layer to display on map',
'raster:bands': [{'scale': 1.0,
'nodata': -1.2676506002282294e+30,
'offset': 0.0,
'sampling': 'area',
'data_type': 'float32',
'histogram': {'max': 11639639471292416,
'min': -4024247454269440.0,
'count': 11,
'buckets': [24, 1128, 403168, 64788, 5583, 1096, 252, 64, 19, 4]},
'statistics': {'mean': 343251029873510.25,
'stddev': 563904505347325.8,
'maximum': 11639639471292416,
'minimum': -4024247454269440.0,
'valid_percent': 90.81382751464844}}],
'proj:geometry': {'type': 'Polygon',
'coordinates': [[[-180.0, -90.0],
[180.0, -90.0],
[180.0, 90.0],
[-180.0, 90.0],
[-180.0, -90.0]]]},
'proj:projjson': {'id': {'code': 4326, 'authority': 'EPSG'},
'name': 'WGS 84',
'type': 'GeographicCRS',
'datum': {'name': 'World Geodetic System 1984',
'type': 'GeodeticReferenceFrame',
'ellipsoid': {'name': 'WGS 84',
'semi_major_axis': 6378137,
'inverse_flattening': 298.257223563}},
'$schema': 'https://proj.org/schemas/v0.4/projjson.schema.json',
'coordinate_system': {'axis': [{'name': 'Geodetic latitude',
'unit': 'degree',
'direction': 'north',
'abbreviation': 'Lat'},
{'name': 'Geodetic longitude',
'unit': 'degree',
'direction': 'east',
'abbreviation': 'Lon'}],
'subtype': 'ellipsoidal'}},
'proj:transform': [0.1, 0.0, -180.0, 0.0, -0.1, 90.0, 0.0, 0.0, 1.0]},
'rendered_preview': {'title': 'Rendered preview',
'href': 'https://openveda.cloud/api/raster/collections/no2-monthly/items/OMI_trno2_0.10x0.10_202309_Col3_V4.nc/preview.png?bidx=1&assets=cog_default&rescale=0%2C15000000000000000&resampling=bilinear&color_formula=gamma+r+1.05&colormap_name=rdbu_r',
'rel': 'preview',
'roles': ['overview'],
'type': 'image/png'}},
'geometry': {'type': 'Polygon',
'coordinates': [[[-180, -90],
[180, -90],
[180, 90],
[-180, 90],
[-180, -90]]]},
'collection': 'no2-monthly',
'properties': {'end_datetime': '2023-09-30T00:00:00+00:00',
'start_datetime': '2023-09-01T00:00:00+00:00'},
'stac_version': '1.0.0',
'stac_extensions': ['https://stac-extensions.github.io/raster/v1.1.0/schema.json',
'https://stac-extensions.github.io/projection/v1.1.0/schema.json']}Define an area of interest
We will be using a bounding box over metropolitan france. We’ll use that bounding box to subset the data when calculating the timeseries.
france_bounding_box = {
"type": "Feature",
"properties": {"ADMIN": "France", "ISO_A3": "FRA"},
"geometry": {
"type": "Polygon",
"coordinates": [
[
[-5.183429, 42.332925],
[8.233998, 42.332925],
[8.233998, 51.066135],
[-5.183429, 51.066135],
[-5.183429, 42.332925],
]
],
},
}Let’s take a look at that box.
m = folium.Map(
location=[40, 0],
zoom_start=3,
)
folium.GeoJson(france_bounding_box, name="France").add_to(m)
mUse /statistics to get data for the AOI
First, we create a generate_stats function and then we call it with the bounding box defined for France.
NOTE: The RASTER_API expects AOI as a Feature or a FeatureCollection. The datatype of the input matches the datatype of the output. So if you use a FeatureCollection as input, you will get back a FeatureCollection.
def generate_stats(item, geojson):
""" Generate statistics for a particular item and AOI
NOTE: This function assumes that the AOI is a geojson `Feature`.
"""
response = requests.post(
f"{RASTER_API_URL}/collections/{collection_id}/items/{item['id']}/statistics",
json=geojson
)
assert response.ok, response.text
return {
**response.json()["properties"]["statistics"]["cog_default_b1"],
"start_datetime": item["properties"]["start_datetime"],
}Let’s run this function on a single item to get a sense of the output
%%time
generate_stats(items[0], france_bounding_box)CPU times: user 4.1 ms, sys: 0 ns, total: 4.1 ms
Wall time: 518 ms{'min': -387779578036224.0,
'max': 5971174383157248.0,
'mean': 1636077207936257.2,
'count': 11880.0,
'sum': 1.9436597230282736e+19,
'std': 808762440624020.6,
'median': 1506456557846528.0,
'majority': 1143241374171136.0,
'minority': -387779578036224.0,
'unique': 11875.0,
'histogram': [[120, 1616, 4274, 3441, 1384, 624, 244, 125, 40, 12],
[-387779578036224.0,
248115831504896.0,
884011241046016.0,
1519906650587136.0,
2155802060128256.0,
2791697603887104.0,
3427592744992768.0,
4063488422969344.0,
4699383564075008.0,
5335279242051584.0,
5971174383157248.0]],
'valid_percent': 100.0,
'masked_pixels': 0.0,
'valid_pixels': 11880.0,
'percentile_2': 395780397465600.0,
'percentile_98': 3847018581590016.0,
'start_datetime': '2023-09-01T00:00:00+00:00'}Generate stats
This may take some time due to the complexity of the shape we’re requesting. See the end of this notebook for tips on how to speed this up.
%%time
stats = [generate_stats(item, france_bounding_box) for item in items]CPU times: user 282 ms, sys: 43.8 ms, total: 326 ms
Wall time: 50 sPlot timeseries
It is easier to interact with these results as a pandas dataframe so create one from the list of dicts.
df = pd.DataFrame(stats)Convert the start_datetime column to real pandas datetime dtype:
df["date"] = pd.to_datetime(df["start_datetime"])Construct the plot
fig = plt.figure(figsize=(20, 10))
plt.plot(df["date"], df["mean"], "black", label="Mean monthly NO2 values")
plt.fill_between(
df["date"],
df["mean"] + df["std"],
df["mean"] - df["std"],
facecolor="lightgray",
interpolate=False,
label="+/- one standard devation",
)
plt.plot(
df["date"],
df["min"],
color="blue",
linestyle="-",
linewidth=0.5,
label="Min monthly NO2 values",
)
plt.plot(
df["date"],
df["max"],
color="red",
linestyle="-",
linewidth=0.5,
label="Max monhtly NO2 values",
)
plt.legend()
plt.title("NO2 Values in France (2016-2022)")Text(0.5, 1.0, 'NO2 Values in France (2016-2022)')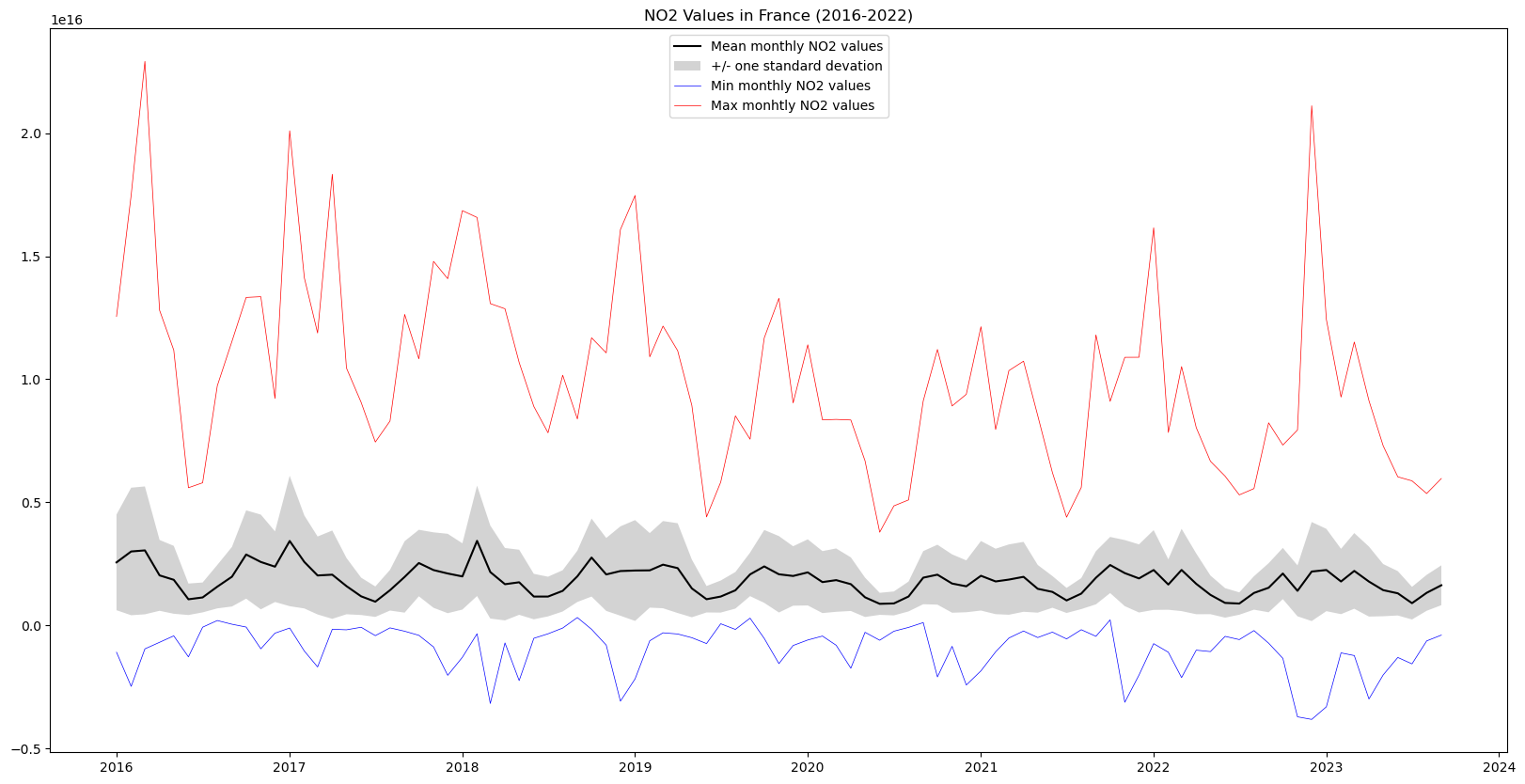
In this graph we can see the yearly cycles in NO2 values due to seasonal variations, as well as a slight downward slope in maximum NO2 values
Complex AOI
The values plotted above don’t correspond exactly to Fance, since the bounding box excludes Corsica and overseas territories such as Mayotte and French Polynesia, and covers parts of neighboring countries including Spain, Italy, Germany and the entirety of Luxembourg. We can fetch GeoJSON from an authoritative online source (https://gadm.org/download_country.html).
While the NO2 values above correspond more or less to those of in France, we can be much more precise by using a complex geojson that represents the boundaries of France exactly, including overseas territories in the Carribean and Indian Oceans, and South America.
Note: In this notebook we write out the whole perimeter as a MultiPolygon in geojson. In practice you will often be reading this kind of shape from a file (usually with the help of geopandas).
response = requests.get(
"https://github.com/wmgeolab/geoBoundaries/raw/9469f09/releaseData/gbOpen/FRA/ADM0/geoBoundaries-FRA-ADM0.geojson"
)
# If anything goes wrong with this request output error contents
assert response.ok, response.text
result = response.json()
print(f"There are {len(result['features'])} features in this collection")There are 1 features in this collectionThat is the geojson for a feature collection, but since there is only one feature in it we can grab just that.
france_aoi = result["features"][0]Let’s take a look at this AOI on a map
m = folium.Map(
location=[46, 0],
zoom_start=5,
)
folium.GeoJson(france_aoi, name="France").add_to(m)
mSimplifying the AOI
You might notice that that is a highly detailed border. In fact it is so detailed that it is too big to pass to the Raster API. It will get rejected. The limit on the Raster API is 16 Kb. And our current AOI is roughly:
size = sys.getsizeof(json.dumps(france_aoi))
print(f"{size // 1024} Kb")137 KbSo can we simplify it? Naively let’s try just using shapely’s .simplify method:
france = shapely.from_geojson(json.dumps(france_aoi))
france_simplified = france.simplify(0.01)
size = sys.getsizeof(shapely.to_geojson(france_simplified))
print(f"{size // 1024} Kb")30 KbStill too big… what if we increase the tolerance? Basically you can increase it as high as you want but you’ll never get below the 16 Kb threshold because there are a ton of little islands. What if we applied a buffer and then unapplied it before simplifying:
france_simplified = (
france
.buffer(0.1).buffer(-0.1)
.simplify(0.01)
)
size = sys.getsizeof(shapely.to_geojson(france_simplified))
print(f"{size // 1024} Kb")17 KbThat is getting better, but if we look at the coordinates we can see that they have a ton of precision that is unnecessary:
shapely.to_geojson(france_simplified)[:100]'{"type":"MultiPolygon","coordinates":[[[[8.542647846187508,42.232268053459805],[8.561307373976677,42'Maybe we can round all of those.
france_simplified = shapely.set_precision(
france
.buffer(0.1).buffer(-0.1)
.simplify(0.01),
0.0001
)
size = sys.getsizeof(shapely.to_geojson(france_simplified))
print(f"{size // 1024} Kb")7 KbLet’s make sure that AOI still looks decent:
m = folium.Map(
location=[46, 0],
zoom_start=5,
)
folium.GeoJson(france_simplified, name="France").add_to(m)
mWe can now request the NO2 values for this AOI the same way as for the bounding box.
Notice, however, that due to the complexity of the shape (even the simplified shape), it takes a little longer to gather the requested data as for the bounding box example above.
france_new_aoi = {
**france_aoi,
"geometry": json.loads(shapely.to_geojson(france_simplified))
}%%time
stats = [generate_stats(item, france_new_aoi) for item in items]CPU times: user 336 ms, sys: 52.7 ms, total: 389 ms
Wall time: 36.7 saoi_df = pd.DataFrame(stats)
aoi_df["date"] = pd.to_datetime(aoi_df["start_datetime"])We can compare the mean monthly NO2 values calculated when using the bounding box and when using the country’s exact borders
fig = plt.figure(figsize=(20, 10))
plt.plot(
df["date"],
df["mean"],
color="blue",
label="Mean monthly NO2 values using bounding box",
)
plt.plot(
aoi_df["date"],
aoi_df["mean"],
color="red",
label="Mean monthly NO2 values using complex AOI",
)
plt.legend()
plt.title("NO2 Values in France (2016-2022)")Text(0.5, 1.0, 'NO2 Values in France (2016-2022)')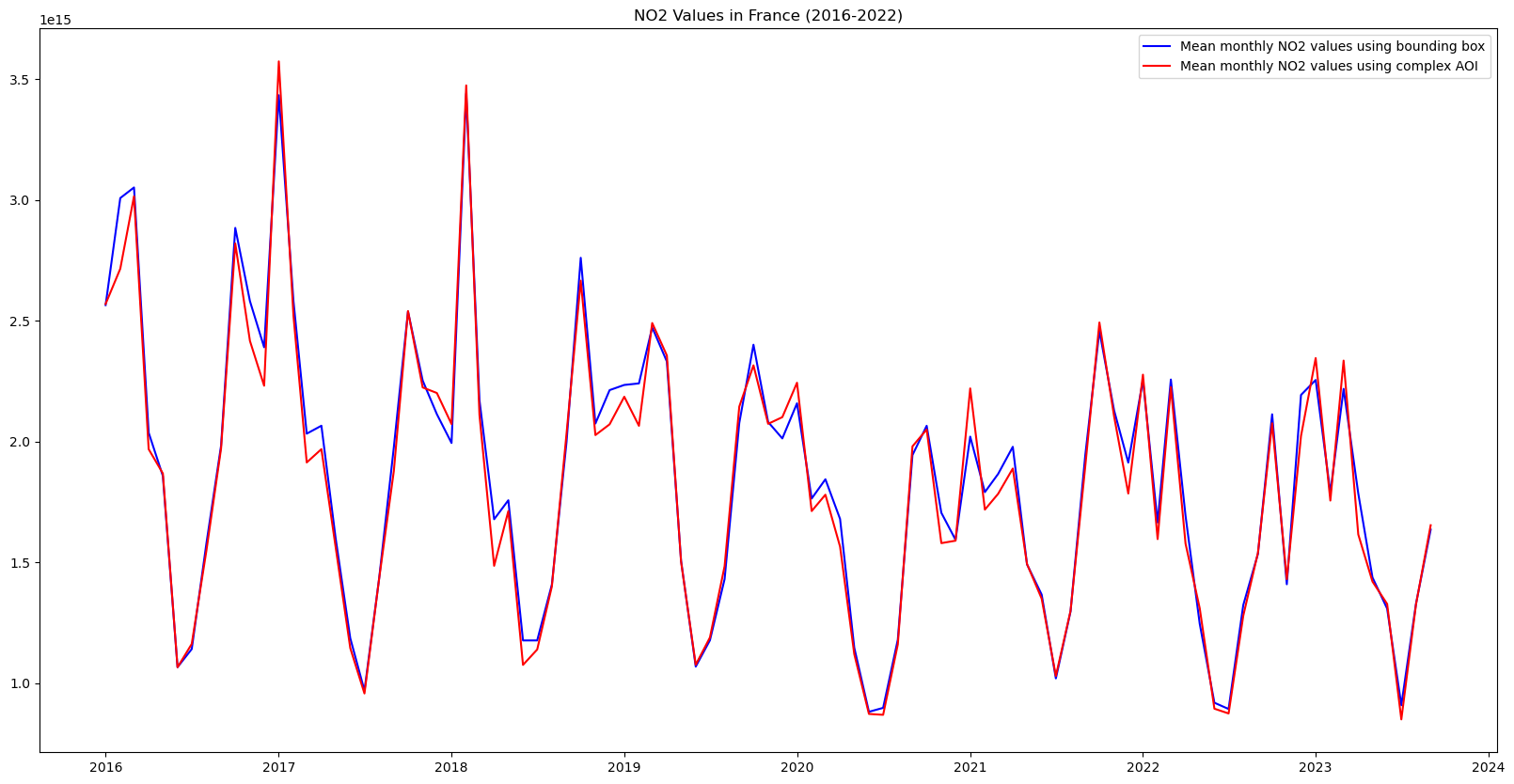
While the difference is small, it is very interesting to note that the NO2 values calculated using the exact borders are systematically less than when using the bounding box. This may be due to the fact that the bounding box includes parts of western Germany and northern Italy that have a lot industrial activity, whereas the areas included when using the exact borders that are not included in the bounding box case, are overseas territories much less industrial activity.
Speed things up: parallelize computation with Dask
We can drastically reduce the time it takes to generate the timeseries, even with the complex AOI above, by parallelizing our code. The /statistics endpoint is powered by AWS Lambda which executes each request in a separate instance. This means the requests are highly scalable. Since each statistics request is for a single timestamp, we can request statistics for multiple timesteps concurrently, and greatly reduce the amount of time needed. We will demonstrate this by using the Dask.
Submit work
First we will create a Dask client. In this case we will use the threads on the same server that is running this jupyter notebook. We will submit the generate_stats function for each item in our list and collect a list of futures. This will immediately kick off work in dask. We can then gather all the results.
%%time
import dask.distributed # noqa
with dask.distributed.Client() as client:
futures = [client.submit(generate_stats, item, france_new_aoi) for item in items]
stats = client.gather(futures)CPU times: user 1.83 s, sys: 126 ms, total: 1.96 s
Wall time: 13.5 sAlternate approach
If you are familiar with the concurrent.futures library you can use that instead of Dask. Or you can use httpx instead of requests to fetch the statistics asynchronously.
